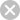Fillable Printable Trifold Poster Template - Utah Valley University
Fillable Printable Trifold Poster Template - Utah Valley University
Trifold Poster Template - Utah Valley University
TEMPLATE DESIGN © 2007
www.PosterPresentations.com
Copper uptake by CMC1 Deletion in
Saccharomyces Cerevisiae
Nick Corbett, Scott Voorhees and Daren Heaton
Utah Valley University, 800 W University Parkway, Orem Utah 84058.
Abstract
Introduction
Materials and Methods Flame Atomic Absorption
Spectroscopy (AAS)
Discussion
References
Fold or cut poster here
Fold or cut poster here
Fold or cut poster here
Fold or cut poster here
Cytochrome c oxidase (CcO) or complex IV is the
terminal component of the electron transport chain. In
eukaryotic organisms, CcO is composed of 12-13
subunits. The core of eukaryotic CcO contains three
mitochondrially encoded subunits that comprise the
catalytic core of the complex and several gene products
encoded for by the nucleus (1). Essential to the redox
function of CcO are several critical cofactors: two
hemes and two copper centers (2). The crystal
structure of CcO has led us to several insights about its
structural components and catalytic activity (3).
However, a large set of nuclear gene products are
essential for CcO activity that are not part of the
structural machinery of the complex (4,5). These
components have been implicated in various stages of
CcO assembly including, heme processing and
insertion (6), Cu
A
and Cu
B
site delivery and insertion
(7), subunit processing and subunit assembly (8,9).
Among them are well characterized CcO assembly
factors that involve the biogenesis of the Cu
A
and Cu
B
sites in CcO. Sco I, Cox17, Cox 11 and Cox23 are all
essential to CcO activity and have properties that
appear to be critical to in the maturation of the Cu
A
and
Cu
B
sites(10-13). Cmc1 has also been implicated in
CcO activity. Cmc1 may have an important role in
copper sorting within the mitochondria (14).
1. Capaldi. R.A. Structure and function of cytochrome c oxidase. Annu. Rev. Biochem 1990, 59:569-596.
2. Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R,
Yaono R, Yoshikawa S. The whole structure of the 13-subunit oxidized cytochrome c oxidase at 2.8 A.
Science 1996 272:1136-44.
3. Tsukihara T, Aoyama H, Yamashita E, Tomizaki T, Yamaguchi H, Shinzawa-Itoh K, Nakashima R,
Yaono R, Yoshikawa S. Structures of metal sites of oxidized bovine heart cytochrome c oxidase at 2.8
A, Science 1995, 269:1069-74.
4. J.E. McEwen, C. Ko, B. Kloeckner-Griussem, R.O. Poyton. Nuclear functions required for
cytochrome c oxidase biogensis in Saccharomyces cerevisiae. Characterization of mutants in 34
complementation groups. J Biol. Chem. 1987 261:11872-11879.
5. A Tzagoloff, C.L. Dieckmann, PET genes of Saccharomyces cerevisiae, Microbiol. Rev. 1990 54:211-
225.
6. Khalimonchuk O, Bestwick M, Meunier B, Watts TC, Winge DR. Formation of the redox cofactor
centers during Cox1 maturation in yeast cytochrome oxidase. Mol Cell Biol. 2010 30:1004-17.
7. Horng Y.C., Cobine P.A., Maxfield A.B., Carr H.S., Winge D.R. Specific copper transfer from the
Cox17 metallochaperone to both Sco1 and Cox11 in the assembly of yeast cytochrome C oxidase. J Biol
Chem. 2004 279:35334-40.
8. Barros MH, Myers AM, Van Driesche S, Tzagoloff A. COX24 codes for a mitochondrial protein
required for processing of the COX1 transcript. J Biol Chem. 2006 281:3743-51.
9. Pierrel F, Khalimonchuk O, Cobine PA, Bestwick M, Winge DR. Coa2 is an assembly factor for yeast
cytochrome c oxidase biogenesis that facilitates the maturation of Cox1. Mol Cell Biol. 2008 16:4927-39.
10.Rigby K, Cobine PA, Khalimonchuk O, Winge DR. Mapping the functional interaction of Sco1 and
Cox2 in cytochrome oxidase biogenesis. J Biol Chem. 2008 283:15015-22.
11.Glerum, D. M., Shtanko, A., and Tzagoloff, A. SCO1 and SCO2 Act as High Copy Suppressors of a
Mitochondrial Copper Recruitment Defect in Saccharomyces cerevisiae J. Biol. Chem. 1996 271:20531–
20535.
12.Barros MH, Johnson A, Tzagoloff A. COX23, a homologue of COX17, is required for cytochrome
oxidase assembly. J Biol Chem. 2004 279:31943-7.
13.Carr HS, Maxfield AB, Horng YC, Winge DR. Functional analysis of the domains in Cox11. J Biol
Chem. 2005 280:22664-9.
14.Darryl Horn, Hassan Al-Ali and Antoni Barrientos. Cmc1p is a Conserved Mitochondrial Twin
CX
9
C Protein Involved in Cytochrome c Oxidase Biogenesis. Mol. Cell. Biol. 2008 28:4354-4363
15.Crouch S, Holler, F. Skoog, D. (2007). Principles of Instrumental Analysis 6
th
Edition. Belmont: Thomson
Brooks/Cole. 230-250.
The CMC1 deletions appears to uptake less copper
when media is supplement with .04% copper
sulfate or higher.
Results
MATERIALS AND METHODS
A 20% glucose solution and minimal media solution (5 g of
ammonium sulfate, 1.7 g of yeast nitrogen base without
amino acids, 0.73 g of amino acid dropout mix (CSM-ADE-
TRP)) were made. Poured 45 mL of minimal media into
separate 250 mL Erlenmeyer flasks and then autoclaved the
minimal media and the 20% glucose solutions. After the
solutions cooled down, added 5 mL of the 20% glucose
solution to the Erlenmeyer flasks.
Inoculated one of the 250 mL Erlenmeyer flasks with the
wild type cells and another flask with the CMC1 cells and
allowed them to grow for less than 24 hours in a 30°C
incubator rotating at 250 rpm. Once OD reached 0.3 (1:10
dilution)—UV-Vis spectrometer measurement—transferred
the calculated amount of cells to the shock series flasks and
allowed to grow for less than 24 hours. Once OD reached
0.3 (1:10 dilution)—UV-Vis spectrometer measurement—
from the shock series flasks, the corresponding copper
percent was added and the shock series started with 15
minute 1 mL samples taken for up to two hours (0.5 M Cu
solution was prepared from 19.95 g of CuSO
4
in a 250 mL
volumetric flask).
The 1 mL samples were centrifuged in 2 mL micro
centrifuge tubes twice with the second centrifuge done with
the removal of the supernatant. Removed 0.5 mL of these
different cell solutions, added them to separate appropriately
labeled 50 mL volumetric flasks, and then filled to the mark
of the flask with 2% nitric acid. Transferred these solutions
to appropriately labeled 50 mL conical tubes to be run on the
AAS.
Made 2% nitric acid solutions and made 0.5 ppm, 1 ppm, 1.5
ppm, 2 ppm, 2.5 ppm, and 3 ppm Cu standards (The blank
was the 2% nitric acid solution with no Cu added). After
calibrating the AAS, the samples were run.
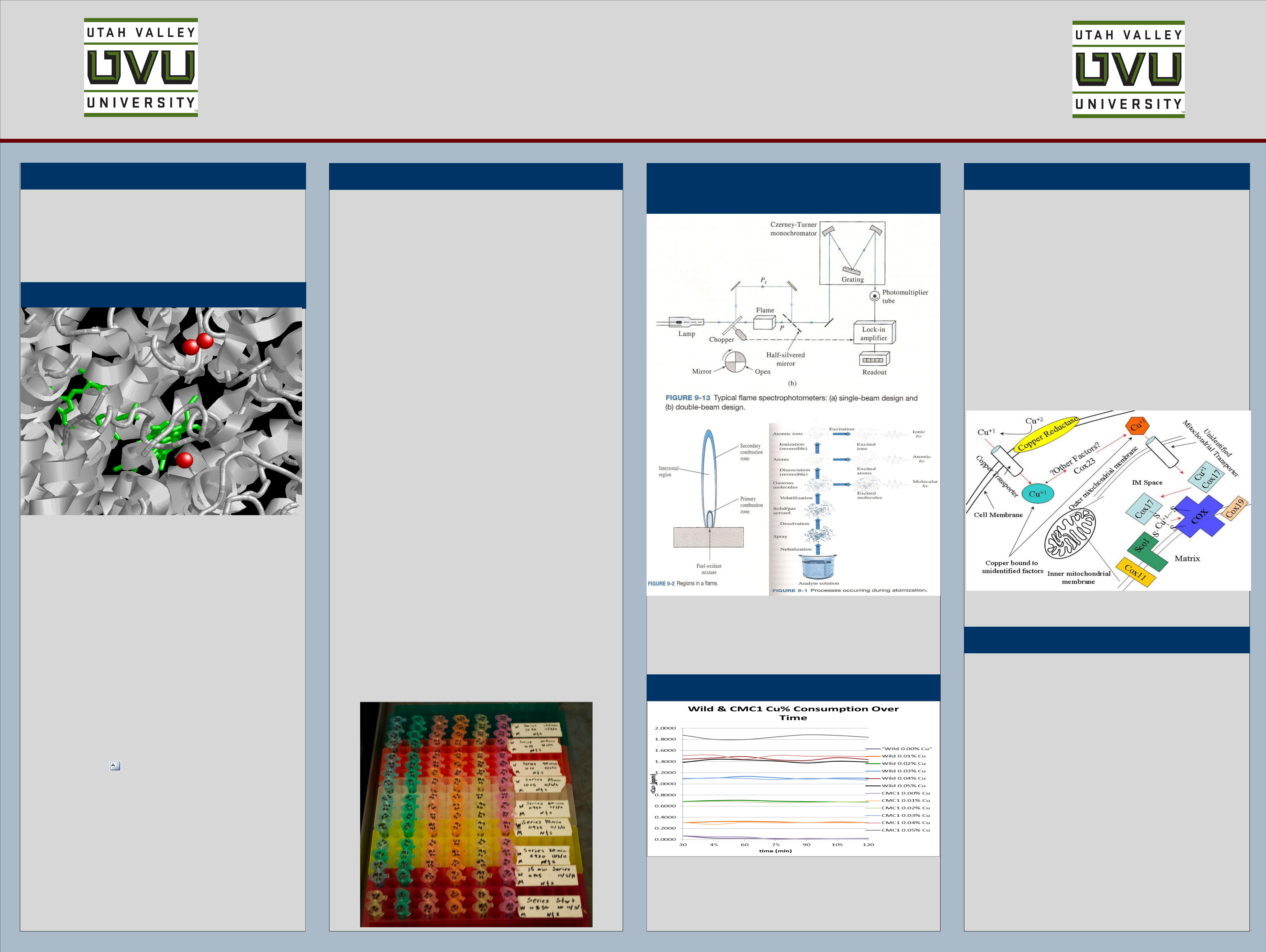
Discrepancies in the amount of Cu uptake between the two (CMC1 &
Wild), over the 2hr time period, was only observed at the upper limit
of the Cu% (0.04% and 0.05% Cu) added. Though the 0.04% Cu
series only showed minor differences in the amount of Cu uptake the
trend drastically increased at the 0.05% Cu series.
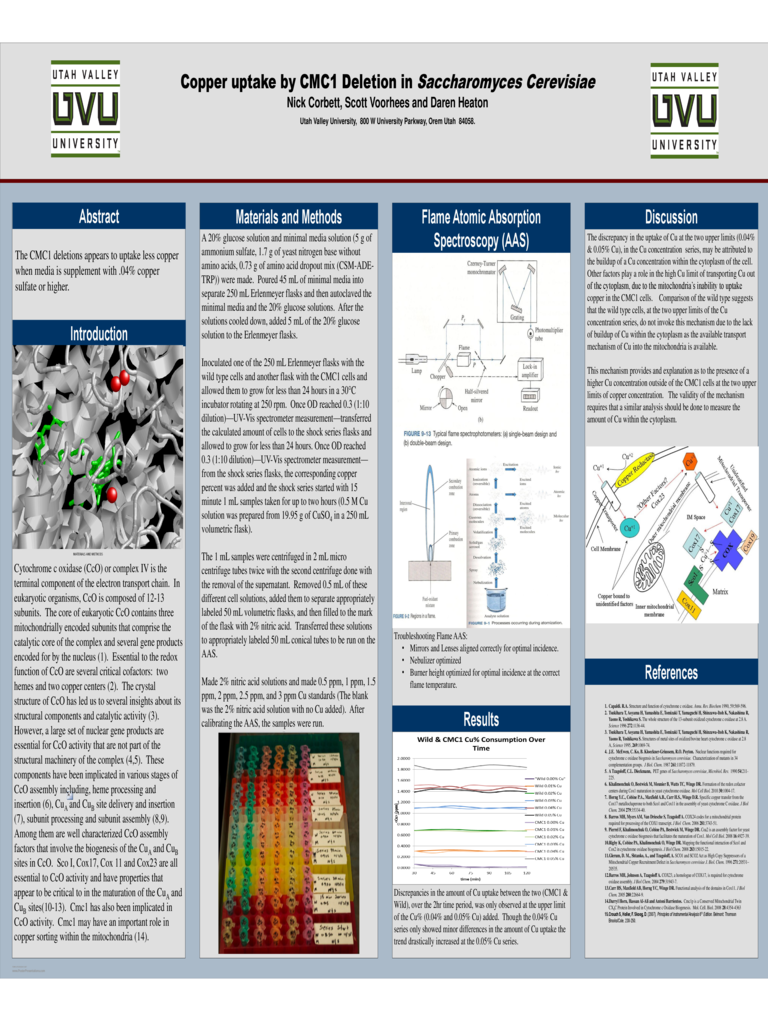
Troubleshooting Flame AAS:
• Mirrors and Lenses aligned correctly for optimal incidence.
• Nebulizer optimized
• Burner height optimized for optimal incidence at the correct
flame temperature.
The discrepancy in the uptake of Cu at the two upper limits (0.04%
& 0.05% Cu), in the Cu concentration series, may be attributed to
the buildup of a Cu concentration within the cytoplasm of the cell.
Other factors play a role in the high Cu limit of transporting Cu out
of the cytoplasm, due to the mitochondria’s inability to uptake
copper in the CMC1 cells. Comparison of the wild type suggests
that the wild type cells, at the two upper limits of the Cu
concentration series, do not invoke this mechanism due to the lack
of buildup of Cu within the cytoplasm as the available transport
mechanism of Cu into the mitochondria is available.
This mechanism provides and explanation as to the presence of a
higher Cu concentration outside of the CMC1 cells at the two upper
limits of copper concentration. The validity of the mechanism
requires that a similar analysis should be done to measure the
amount of Cu within the cytoplasm.